DNA Transposon Termini Signatures (version 1.1)
The termini of most Class II DNA transposons, in particular those with terminal inverted repeat sequences (TIRs), are critical to the process of transposition and often show signs of conservation between families within a single class. These conserved patterns may be used to classify de-novo identified elements. Non-coding DNA transposons often can only be classified by recognition of these patterns along with not-always-available target site duplication characteristics. As many transposons have (fragments) of other elements embedded, even those with apparent coding sequence should generally be classified by their terminal patterns if there is a conflict.
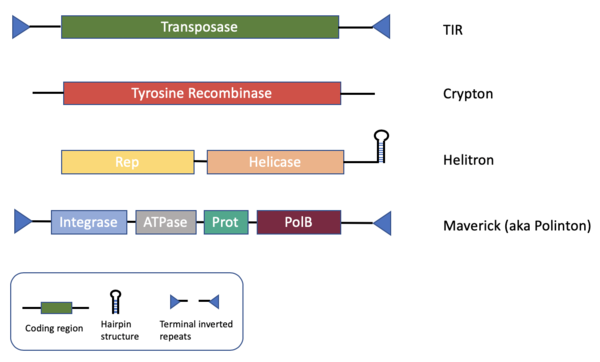
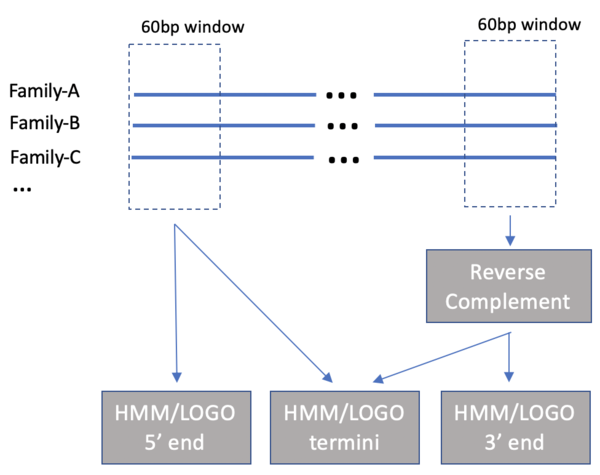
DNA transposons found in eukaryotic genomes fall into four broad groups1,2,3 (Figure 1) each with their own set of autonomous and non-autonomous forms (typically non-coding deletion products retaining the end recognition sites). Here we provide logos and HMMs for the terminal 26 bp of most currently recognized subclasses. The strongest signals are carried by TIR DNA transposons, but even Helitrons and some Crypton elements lacking TIRs contain signs of base conservation. HMMs for the 5’ and 3’ ends were generated from an ungapped alignment of the first/last 60bp of all known members with clearly defined termini (Figure 2). Minor modifications were made to some consensus sequences to have each line up from the true end, most commonly involving removal of target site duplications. The 3’ ends were reverse-complemented to match the 5’ end (in case of TIRs). HMMs were developed using HMMER (hmmbuild) for the 5’ ends, the 3’ ends, and the combined termini. The logos below are a representation of the information content at each position within the HMM. The full set may be downloaded from https://www.dfam.org/releases/dna_termini_1.1 and will be updated as new families are added to each class.
- Piégu, Benoît, et al. "A survey of transposable element classification systems–a call for a fundamental update to meet the challenge of their diversity and complexity." Molecular phylogenetics and evolution 86 (2015): 90-109.
- Feschotte, Cédric, and Ellen J. Pritham. "DNA transposons and the evolution of eukaryotic genomes." Annu. Rev. Genet. 41 (2007): 331-368.
- Wicker, Thomas, et al. "A unified classification system for eukaryotic transposable elements." Nature Reviews Genetics 8.12 (2007): 973-982.
| Subclass | Families | Termini | 5' end | 3' end | Notes |
|---|---|---|---|---|---|
| Circular dsDNA Intermediate | |||||
| Crypton_A | 60 |  |  |  | |
| Crypton_C | 5 |  |  |  | |
| Crypton_F | 22 |  |  |  | |
| Crypton_H | 7 |  |  |  | |
| Crypton_I | 9 |  |  |  | |
| Crypton_R | 6 |  |  |  | |
| Crypton_S | 59 |  |  |  | |
| Crypton_V | 40 |  |  |  | |
| DNA Polymerase | |||||
| Maverick | 112 |  |  |  | |
| Rolling Circle | |||||
| Helitron | 930 |  |  |  | |
| Terminal Inverted Repeat | |||||
| Academ_1 | 85 |  |  |  | |
| Academ_2 | 3 |  |  |  | |
| Academ_H | 18 |  |  |  | First C is likely part of the target site that favors CC<cut>. |
| CMC_Chapaev | 46 |  |  |  | |
| CMC_Chapaev_3 | 48 |  |  |  | |
| CMC_EnSpm | 609 |  |  |  | |
| CMC_Mirage | 5 |  |  |  | Could be multiple groups here. Too few families to tell yet. |
| CMC_Transib | 118 |  |  |  | |
| Dada | 29 |  |  |  | |
| Ginger | 60 |  |  |  | |
| hAT combined | 3084 |  |  |  | |
| hAT_Ac | 823 |  |  |  | |
| hAT_Blackjack | 130 |  |  |  | |
| hAT_Charlie | 729 |  |  |  | |
| hAT_Pegasus | 7 |  |  |  | |
| hAT_Restless | 4 |  |  |  | |
| hAT_Tag1 | 158 |  |  |  | |
| hAT_Tip100 | 705 |  |  |  | |
| hAT_hAT1 | 6 |  |  |  | |
| hAT_hAT5 | 33 |  |  |  | |
| hAT_hAT6 | 8 |  |  |  | |
| hAT_hAT19 | 81 |  |  |  | |
| hAT_hATm | 140 |  |  |  | |
| hAT_hATw | 15 |  |  |  | |
| hAT_hATx | 26 |  |  |  | |
| hAT_hobo | 40 |  |  |  | |
| IS3EU | 24 |  |  |  | |
| Kolobok_E | 8 |  |  |  | |
| Kolobok_H | 6 |  |  |  | |
| Kolobok_Hydra | 70 |  |  |  | |
| Kolobok_T2 | 153 |  |  |  | |
| MULE_F | 12 |  |  |  | |
| MULE_MuDR | 1485 |  |  |  | |
| MULE_NOF | 25 |  |  |  | |
| Merlin | 79 |  |  |  | |
| Novosib | 8 |  |  |  | |
| P | 188 |  |  |  | |
| P_Fungi | 16 |  |  |  | |
| PIF_Harbinger | 1093 |  |  |  | |
| PIF_HarbS | 21 |  |  |  | Consensus probably starts with GGGC like other Harbingers; the T could be part of a TSD. |
| PIF_ISL2EU | 44 |  |  |  | Low signal in our dataset. Han et al. (PMID26120370) has an alternative LOGO. |
| PIF_Spy | 56 |  |  |  | |
| PiggyBac | 310 |  |  |  | After removal of TTAA TSDs for many families. There appear to be subgroups with CACGTT…, CACTA… and CTAGTGTCTA termini. |
| PiggyBac_A | 4 |  |  |  | |
| PiggyBac_X | 43 |  |  |  | |
| Sola_1 | 119 |  |  |  | |
| Sola_2 | 89 |  |  |  | |
| Sola_3 | 28 |  |  |  | |
| TcMar combined | 2624 |  |  |  | |
| TcMar_Ant1 | 20 |  |  |  | Appears to be two groups present (TA TSDs removed) in this subclass. One with a CAGCTATT motif and another with TTGTGTTT. |
| TcMar_Cweed | 26 |  |  |  | |
| TcMar_Fot1 | 220 |  |  |  | |
| TcMar_Gizmo | 3 |  |  |  | |
| TcMar_ISRm11 | 109 |  |  |  | |
| TcMar_m44 | 45 |  |  |  | |
| TcMar_Mariner | 457 |  |  |  | |
| TcMar_Mogwai | 3 |  |  |  | |
| TcMar_Pogo | 120 |  |  |  | |
| TcMar_Sagan | 34 |  |  |  | |
| TcMar_Stowaway | 152 |  |  |  | The majority (102) start with CTCCCTC.., but the TA may be part of the TIR as many TA...TA consensi are annotated to have TA TSDs. |
| TcMar_Tc1 | 924 |  |  |  | |
| TcMar_Tc2 | 108 |  |  |  | |
| TcMar_Tc4 | 58 |  |  |  | |
| TcMar_Tigger | 310 |  |  |  | |
| Zator | 51 |  |  |  | |
| Zisupton | 22 |  |  |  | |

